Predicting the Likelihood of Molecules to Act as Modulators of Protein–Protein Interactions | Journal of Chemical Information and Modeling
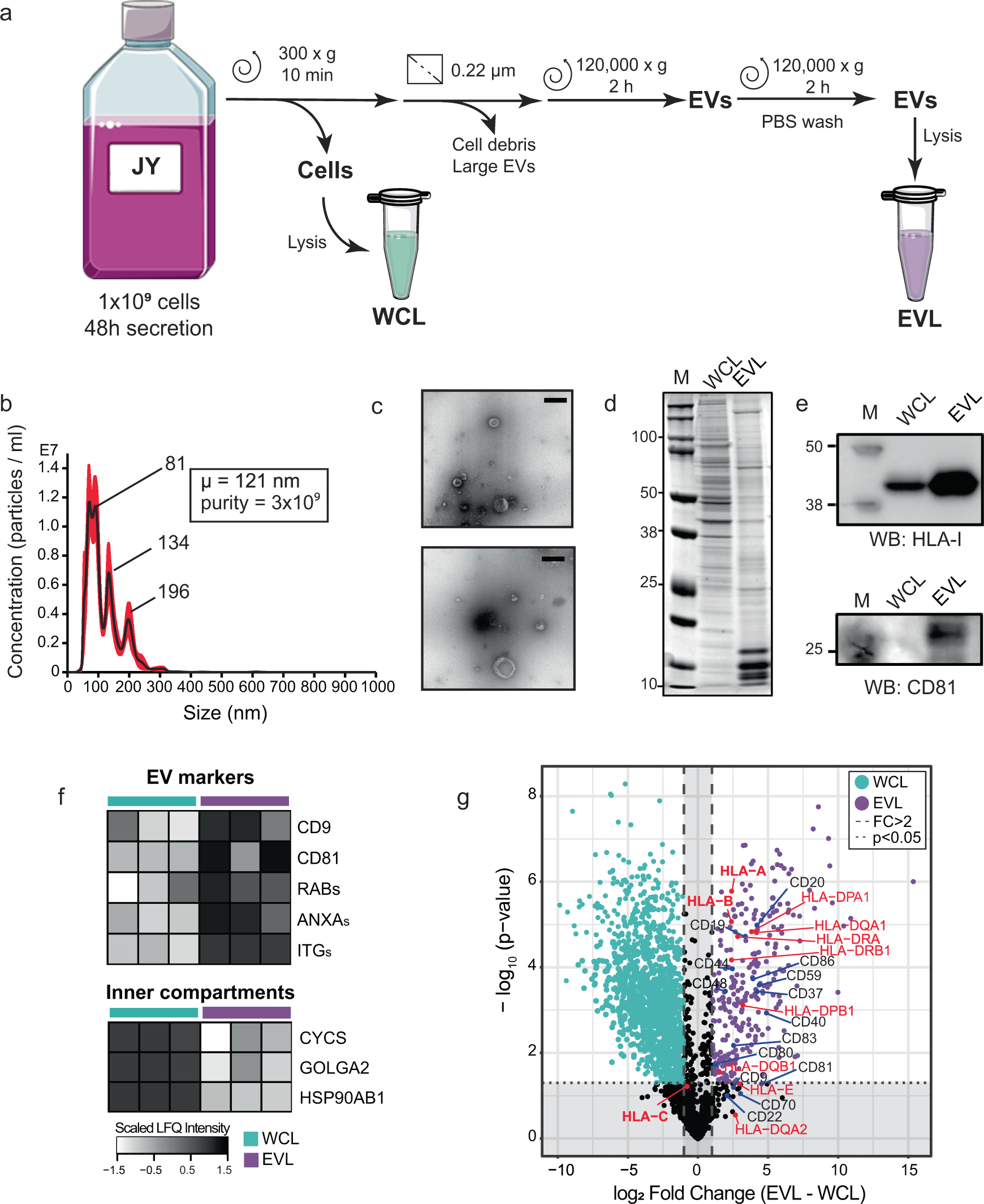
HLA-B and cysteinylated ligands distinguish the antigen presentation landscape of extracellular vesicles | Communications Biology

Modulation of MHC-E transport by viral decoy ligands is required for RhCMV/SIV vaccine efficacy | Science
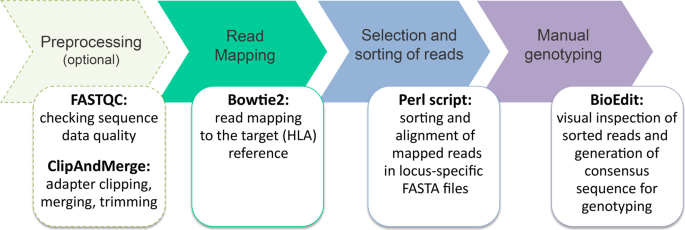
Targeted analysis of polymorphic loci from low-coverage shotgun sequence data allows accurate genotyping of HLA genes in historical human populations | Scientific Reports

Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer - ScienceDirect

Predicting the Affinity of Peptides to Major Histocompatibility Complex Class II by Scoring Molecular Dynamics Simulations | Journal of Chemical Information and Modeling
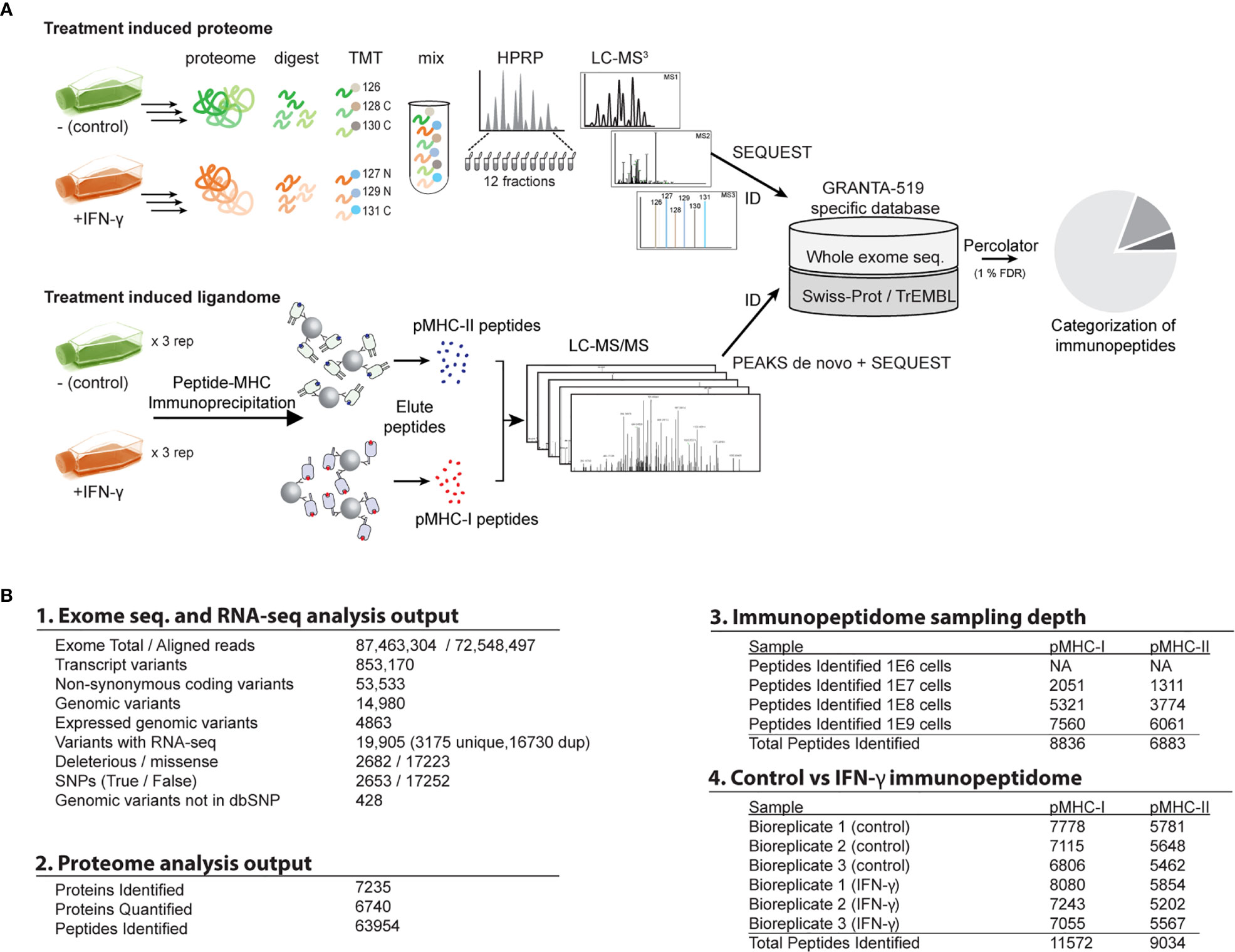
Frontiers | An Integrated Genomic, Proteomic, and Immunopeptidomic Approach to Discover Treatment-Induced Neoantigens
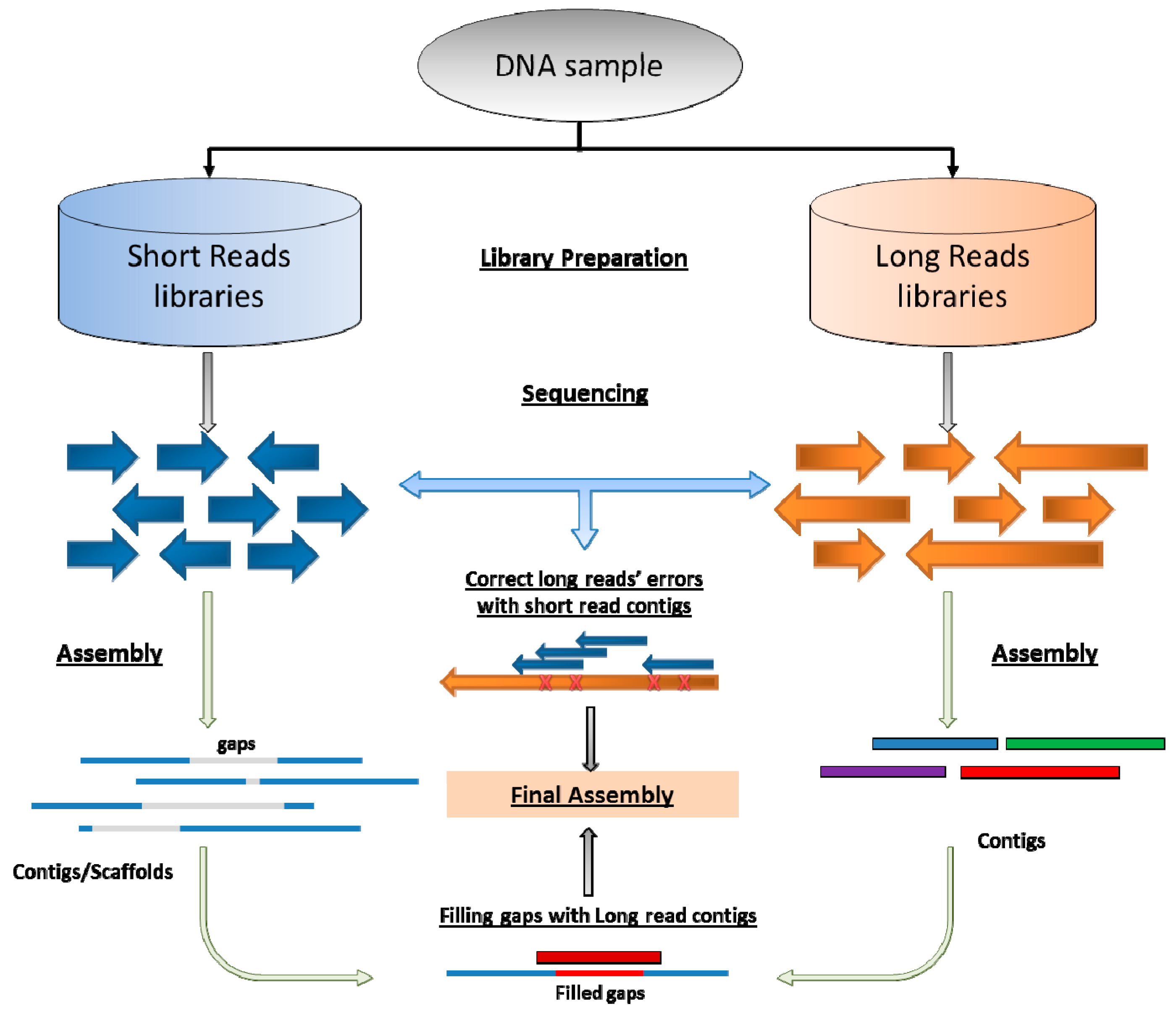
Pharmaceutics | Free Full-Text | Challenges, Solutions, and Quality Metrics of Personal Genome Assembly in Advancing Precision Medicine

Evolutionary origins and interactomes of human, young microproteins and small peptides translated from short open reading frames - ScienceDirect

Systematic genetic analysis of the MHC region reveals mechanistic underpinnings of HLA type associations with disease | eLife

Challenges and opportunities associated with rare-variant pharmacogenomics: Trends in Pharmacological Sciences
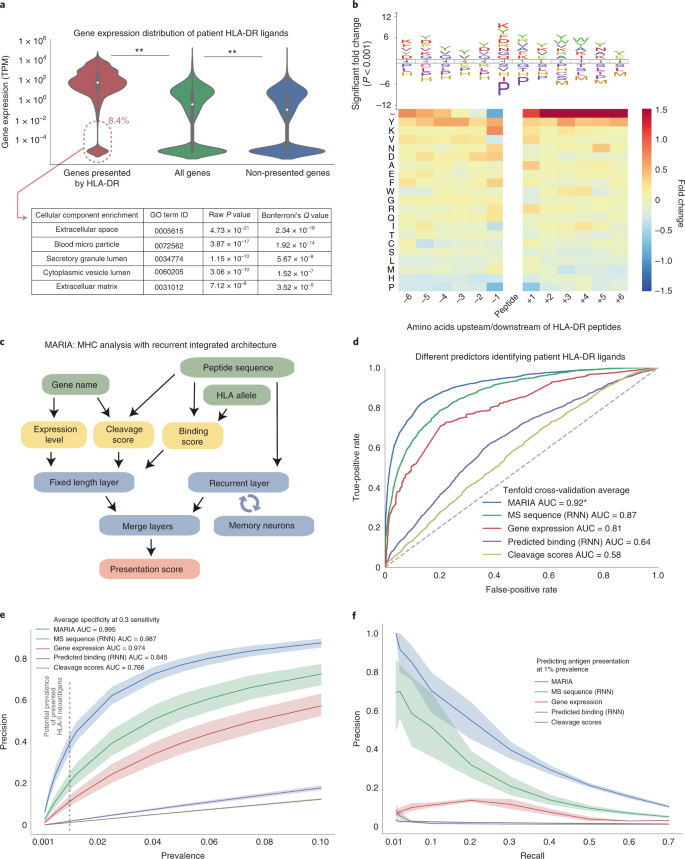
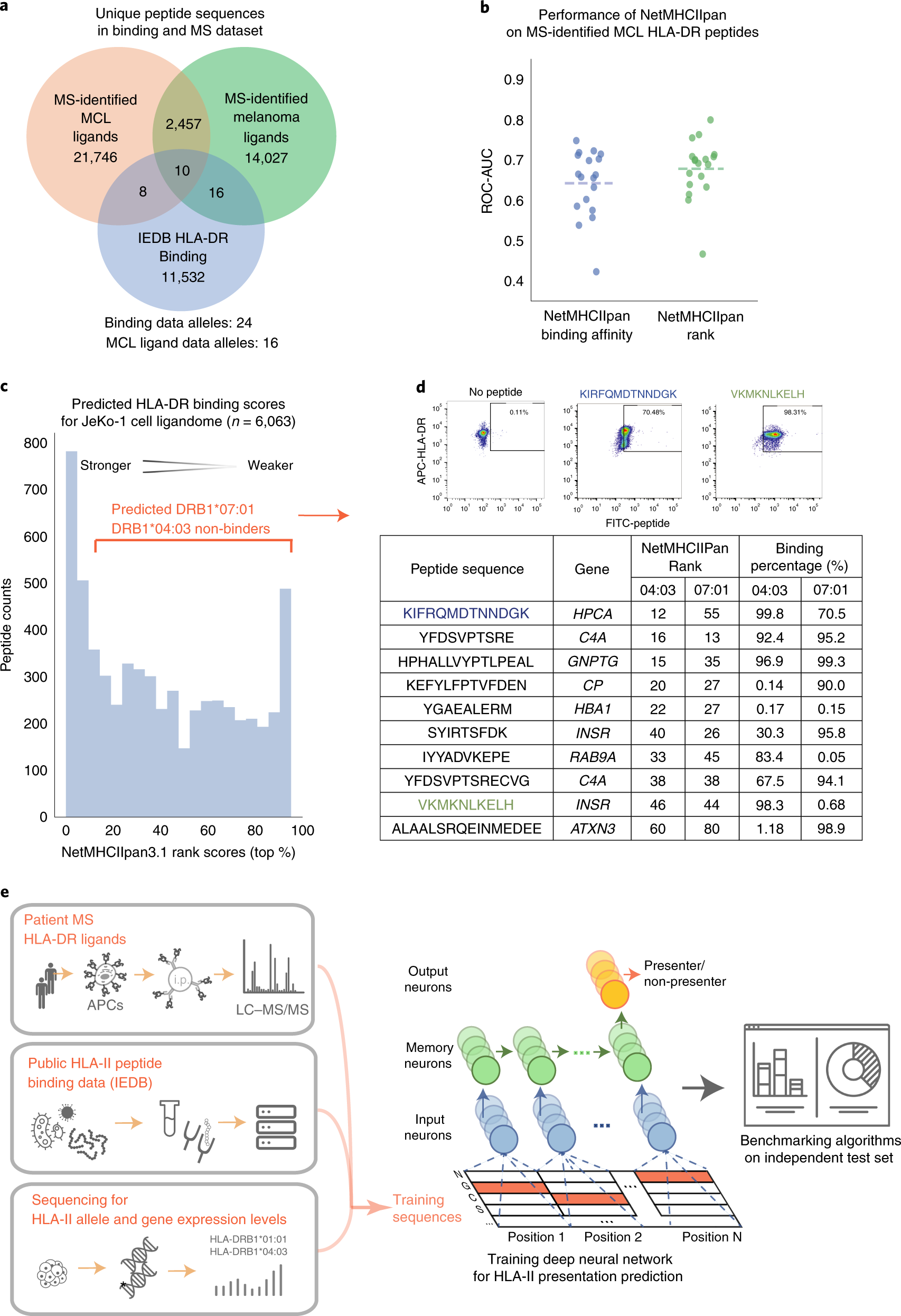
Predicting HLA class II antigen presentation through integrated deep learning | Nature Biotechnology
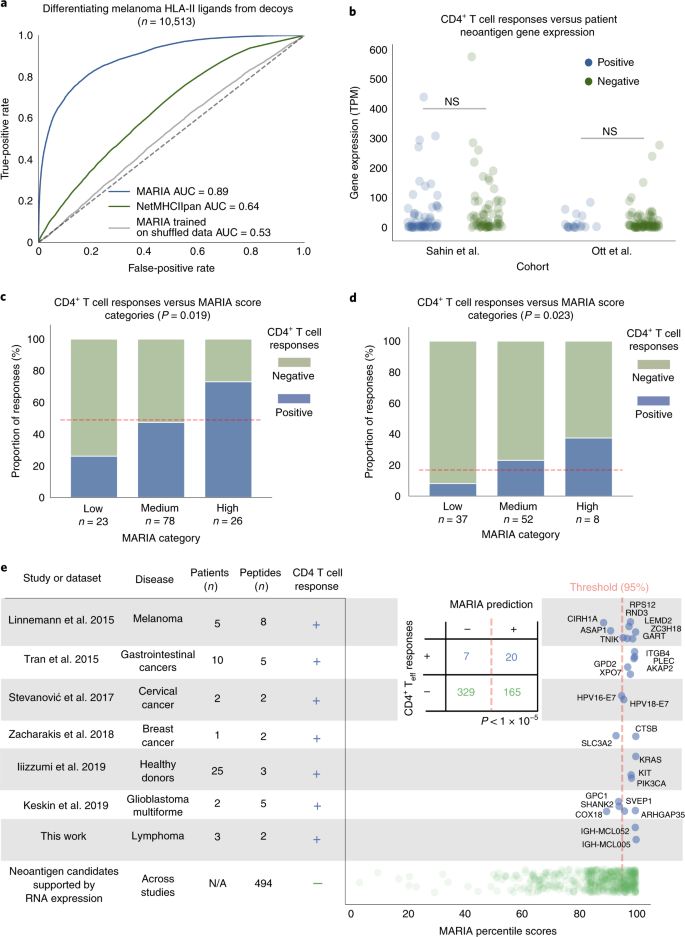
Predicting HLA class II antigen presentation through integrated deep learning | Nature Biotechnology

Deep learning boosts sensitivity of mass spectrometry-based immunopeptidomics | Nature Communications
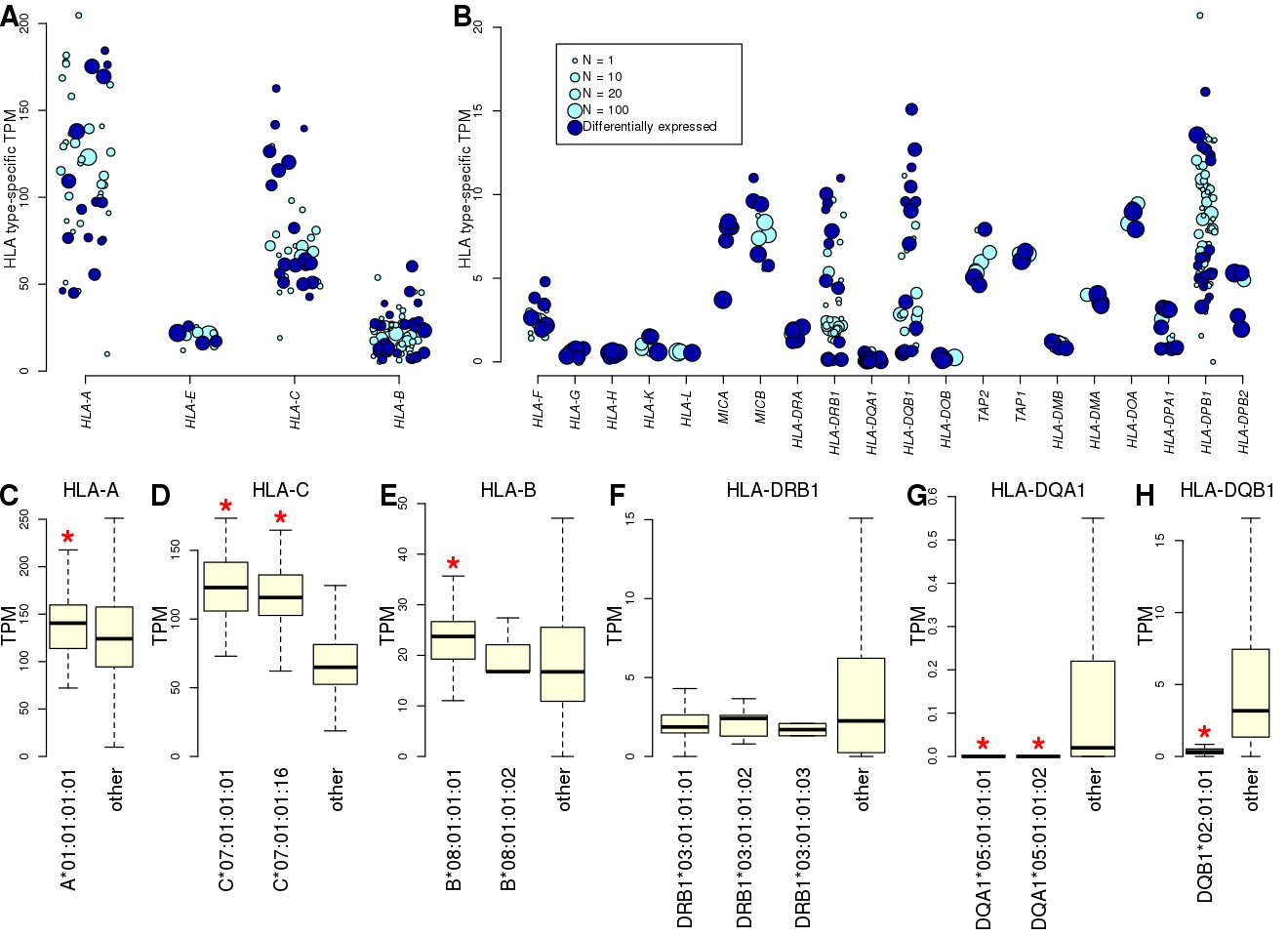
Systematic genetic analysis of the MHC region reveals mechanistic underpinnings of HLA type associations with disease | eLife

Protein-Based Virtual Screening Tools Applied for RNA–Ligand Docking Identify New Binders of the preQ1-Riboswitch | Journal of Chemical Information and Modeling
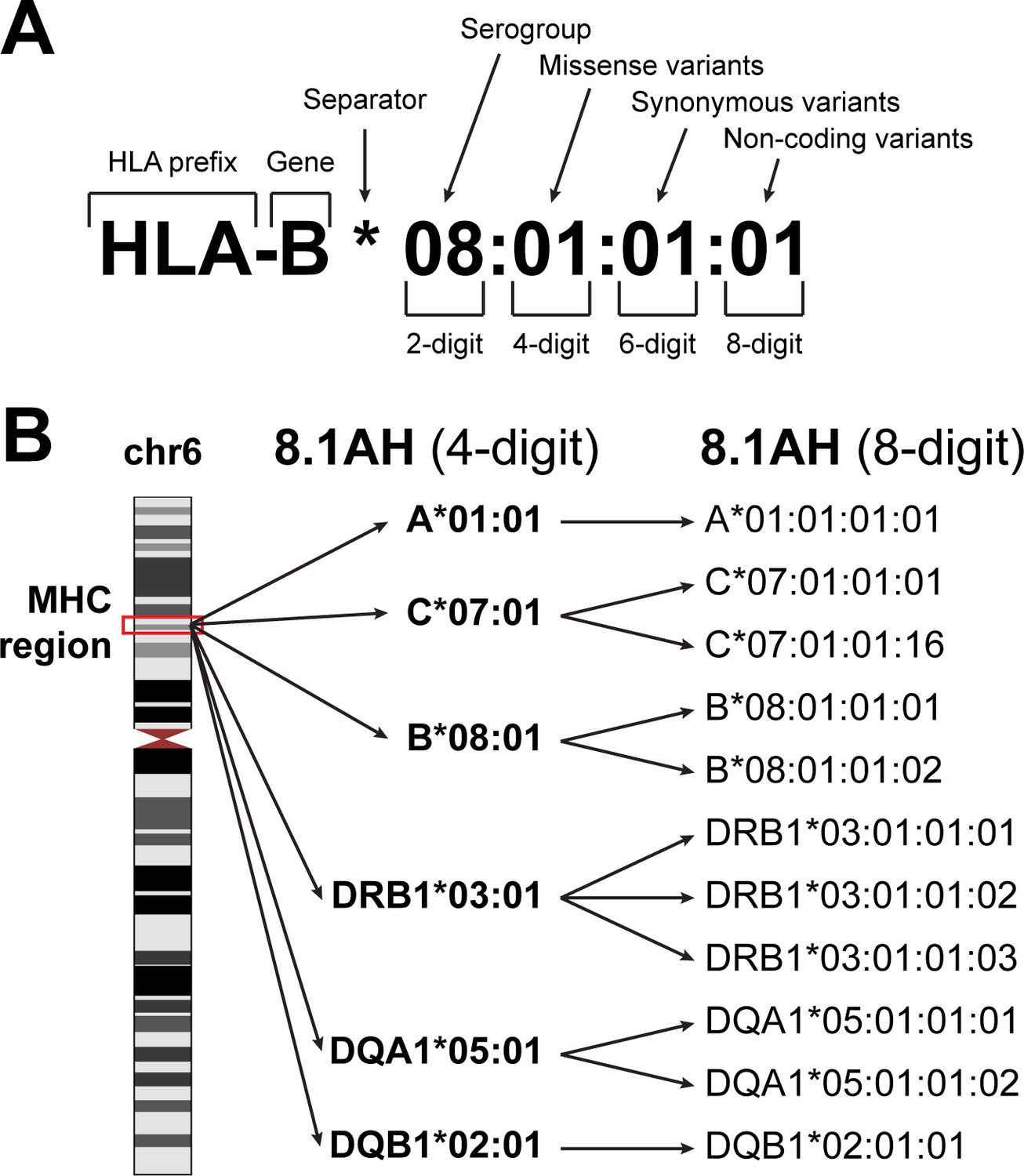
Systematic genetic analysis of the MHC region reveals mechanistic underpinnings of HLA type associations with disease | eLife
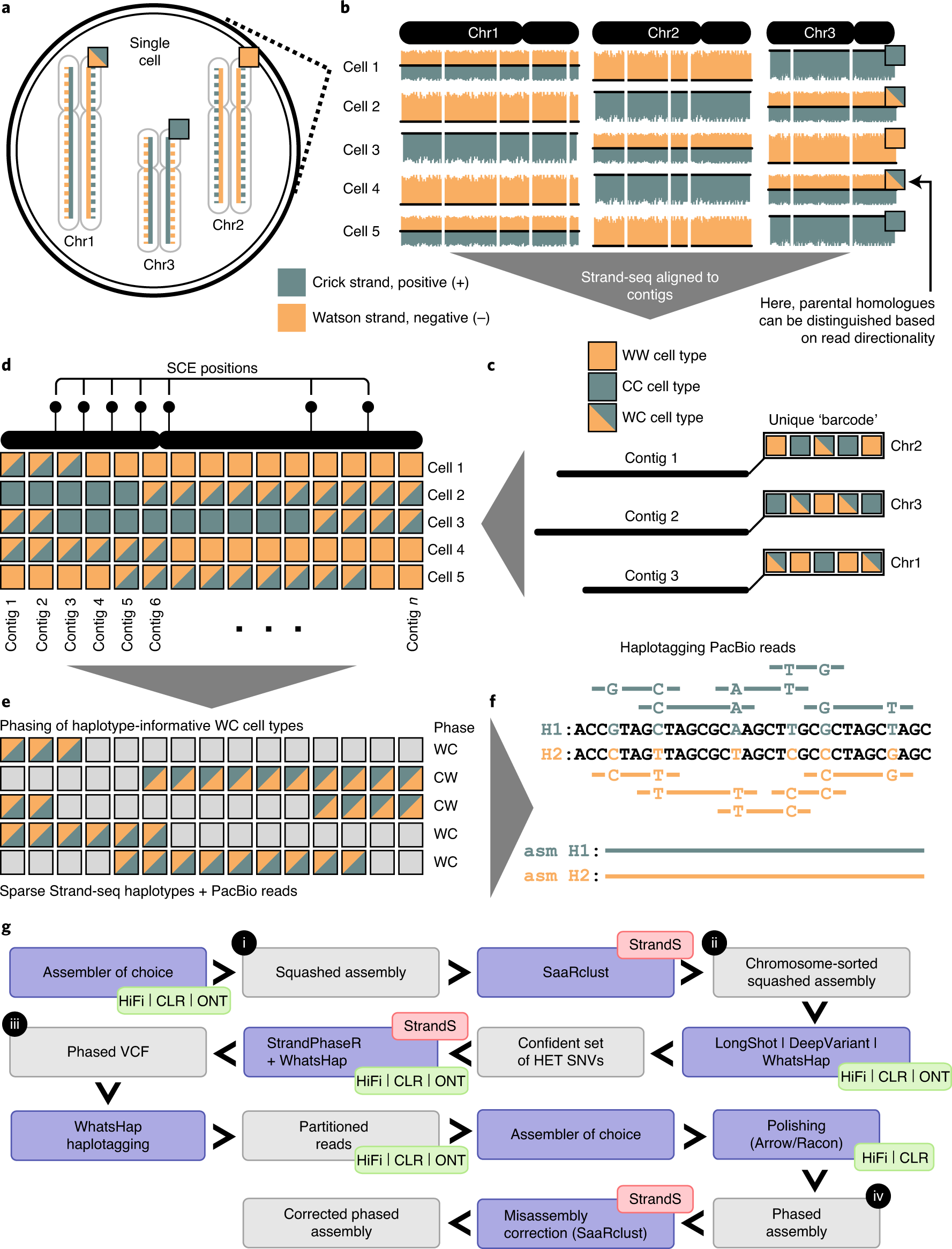
Fully phased human genome assembly without parental data using single-cell strand sequencing and long reads | Nature Biotechnology

Identification of neoantigens in oesophageal adenocarcinoma - Nicholas - Immunology - Wiley Online Library